Time-Resolved Hydroxyl Radical Footprinting
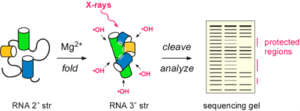
A large experimental challenge is to obtain high-resolution information about RNA structure in a short timespan. Time-resolved hydroxyl radical footprinting provides “snapshots” of RNA tertiary interactions and RNA-protein contacts, at 10-20 millisecond intervals. This method was developed with Michael Brenowitz and Mark Chance at the Center for Synchrotron Biosciences.
In this method, the solvent accessibility of the RNA backbone is determined from its susceptibility to cleavage in the presence of hydroxyl radical. The hydroxyl radicals are generated by 10-30 ms exposures to a synchrotron X-ray beam at Brookhaven National Laboratory, or by Fe(II)-EDTA and hydrogen peroxide for 1-2 ms in a rapid quench apparatus. The fragmentation pattern of the RNA is determined by sequencing. We obtain information about which parts of the RNA have folded in the elapsed interval since the start of the folding reaction.