Small Regulatory RNAs and Sm-Like Proteins
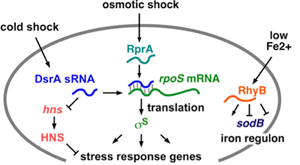
Small regulatory RNAs turn the expression of mRNAs on and off in response to environmental signals. How do they recognize their targets? This is an interesting problem because both the regulator and the mRNA have to refold in order to form the anti-sense complex. Bacterial sRNAs bind an Sm-like protein Hfq, which promotes target recognition and recruits cellular enzymes such as the degradosome and ribosome to the RNA. Sm proteins are found in all kingdoms of life, and are necessary for RNA processing and translation.
We are studying how Hfq protein stimulates antisense pairing between DsrA sRNA and the 5′ leader of rpoS mRNA, using stopped-flow FRET, RNA structure mapping and genetics. The answers will help us understand the function of this important class of RNA-binding proteins, and how enteric bacteria respond to their host environment. A key finding is that mRNAs contains A-rich binding sites for Hfq protein that are required for regulation of the mRNA by sRNAs. We have also found that Hfq forms transient ternary complexes with complementary RNAs, but that Hfq must cycle off the RNA in order for the RNAs to finish base pairing.