RNA Folding During Ribosome Assembly
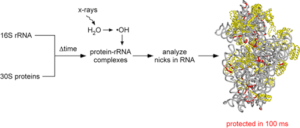
An important problem in molecular biology is how interactions between proteins and newly transcribed RNAs direct the assembly of functional RNPs. We used hydroxyl radical footprinting to follow the emergence of RNA tertiary interactions and RNA-protein interactions during 30S assembly in real time.
Key findings from time-resolved footprinting of the 16S rRNA are that many RNA tertiary interactions present in the 30 S ribosome can form within 5 seconds. However, this process is inefficient because a fraction of the 16 S rRNA population misfolds. As expected, ribosomal proteins stabilize the RNA tertiary structure. However, many proteins protect their rRNA binding sites in stages, with some contacts forming very rapidly, and others taking several seconds or even minutes to be complete. This apparent “induced fit” of RNA-protein binding in the early stages of ribosome assembly may enable the stable addition of other proteins to the complex, or exert “quality control” to ensure that initial RNA interactions are correctly established before subsequent steps in the assembly pathway.