RNA Folding and Dynamics
Ribozymes are excellent models for RNA folding mechanisms. We are studying the folding pathways of group I ribozymes using a combination of biochemical and physical methods such as time-resolved hydroxyl radical footprinting, stopped-flow fluorescence, neutron spectroscopy and small angle X-ray scattering. We have also developed simple reporter assays for evaluating RNA folding in bacteria and yeast.
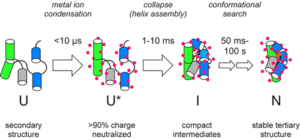
Folding studies have revealed principles of RNA folding that help us understand more complicated processes such as ribosome assembly or RNA regulation. In the presence of cations, RNA double helices assemble into structured and compact intermediates. Subsequent conformational changes lead to the native, catalytically active structure. Tertiary interactions form early during assembly of core helices, and cooperatively stabilize native-like intermediates. Because cooperativity between tertiary interactions in different parts of the RNA favors correct, it improves the accuracy of the folding process.
Stopped flow SAXS experiments show that the initial collapse transition occurs in less than a millisecond. However, depending on the RNA sequence and the folding condition, this collapse can produce incorrect structures that persist for minutes or hours. Refolding of metastable conformations depends strongly on RNA sequence and on ionic conditions, with distributed charge-charge interactions producing systems that are more dynamic. Recently, we have found that molecular crowding comparable to intracellular conditions greatly stabilizes the folded RNA, allowing RNA structures to form in much lower Mg2+ concentrations.