We use computational tools (molecular dynamics simulations and machine learning) to study biomolecule electrostatics and charge transfer in biological systems. These are our current research topics.
Ion channels
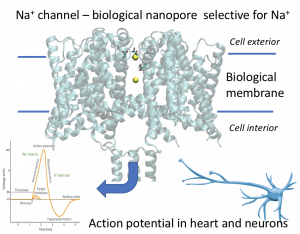 Ion channels are membrane proteins that form pores and control the flow of ions across cell membranes. They are found in virtually all living cells, and have tremendous physiological relevance in regulating ion concentrations inside cells. In the brain and the heart, ion channels are key players in generation of exotic phenomena known as action potentials. Selectivity of ion channels for one ion type is key, as the action potential occurs through rapid opening and closing of voltage dependent sodium selective channels, and potassium selective channels.
Ion channels are membrane proteins that form pores and control the flow of ions across cell membranes. They are found in virtually all living cells, and have tremendous physiological relevance in regulating ion concentrations inside cells. In the brain and the heart, ion channels are key players in generation of exotic phenomena known as action potentials. Selectivity of ion channels for one ion type is key, as the action potential occurs through rapid opening and closing of voltage dependent sodium selective channels, and potassium selective channels.
We currently study the determinants of conductance and selectivity of ion channels. We have recently proposed a new mechanism of how sodium channels prefer sodium vs potassium. If you are interested check out this paper:
Ion channel selectivity through ion-modulated changes of selectivity filter pKa values, PNAS 120 (26), e2220343120
Protein electrostatics
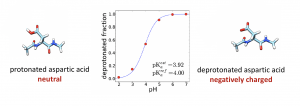 Binding of protons to ionizable amino acids in proteins, such as aspartic and glutamic acid, histidine and lysine affects the charge on that residue and thus determines the protein’s electrostatic properties. Electrostatics affects almost every structural and functional aspect of proteins; including stability, solubility, dynamics, interactions, and catalysis. It is especially important for key processes involving movement of charges, including proton and electron transfer reactions and transport of ions. Thus it is very important to know how many protons are bound to a protein, and to which ionizable residues, i.e., it is important to know what the protonation state of each ionizable residue is.
Binding of protons to ionizable amino acids in proteins, such as aspartic and glutamic acid, histidine and lysine affects the charge on that residue and thus determines the protein’s electrostatic properties. Electrostatics affects almost every structural and functional aspect of proteins; including stability, solubility, dynamics, interactions, and catalysis. It is especially important for key processes involving movement of charges, including proton and electron transfer reactions and transport of ions. Thus it is very important to know how many protons are bound to a protein, and to which ionizable residues, i.e., it is important to know what the protonation state of each ionizable residue is.
We are developing new tools for protein pKa predictions. Our earlier work focused on molecular dynamics based methods for pKa prediction, i.e., on constant pH simulations. Currently we are focusing on machine learning approaches. You can read more about it in our recent paper:
Protein pKa Prediction by Tree-Based Machine Learning, Journal of chemical theory and computation 18 (4), 2673-2686
Membrane electrostatics
Charged lipids play a key role in many cellular processes, including cell migration, endocytosis, exocytosis and signaling. We currently study structural and electrostatic properties of charged lipids, as well as the role of lipid bilayers in sensing of external electric fields.
